Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
About me
This is a page not in th emain menu
Published:
Published:
Published:
A lighthearted project for the geese-averse among us 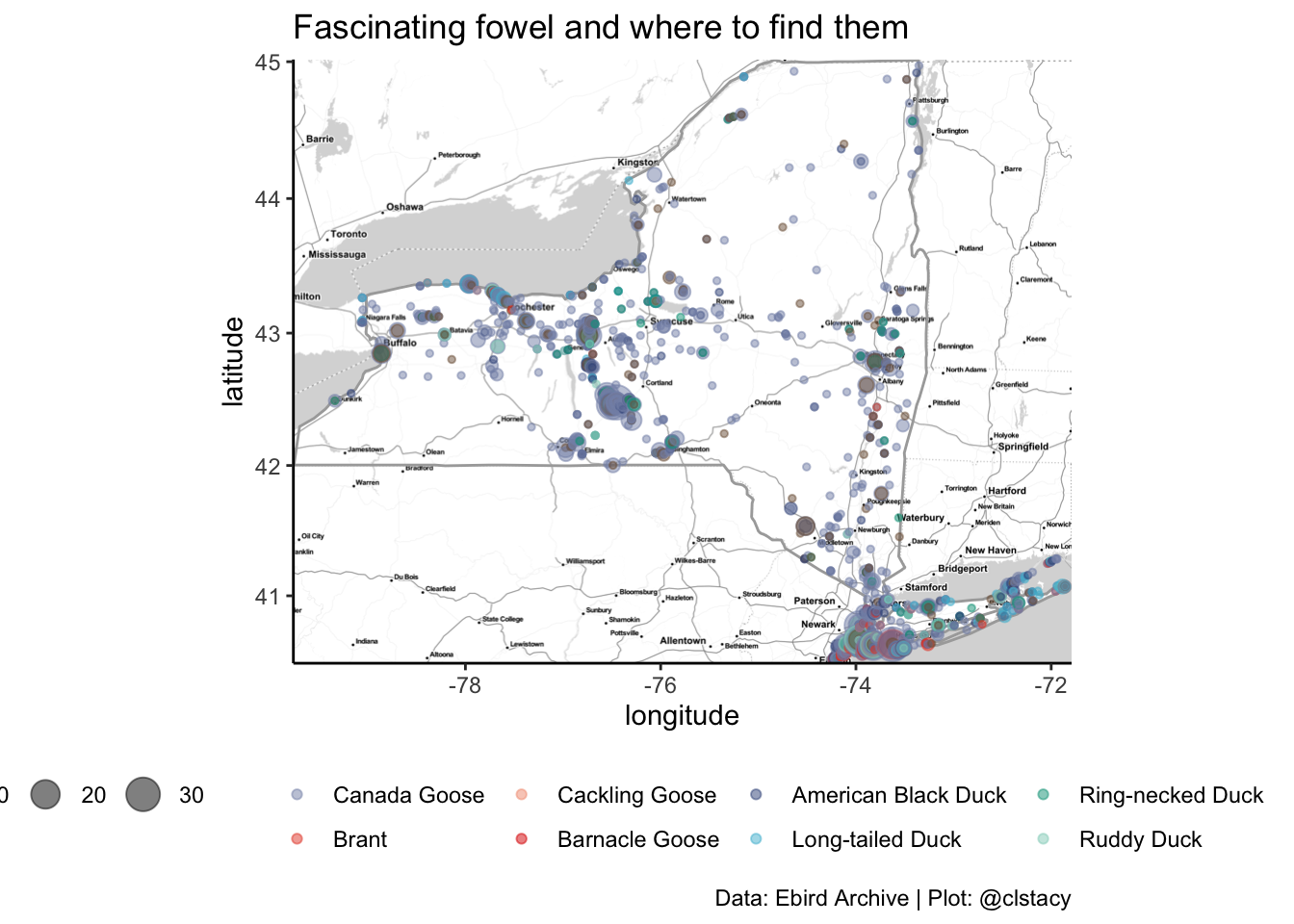
Published:
Course Design in Genomic Data Analysis 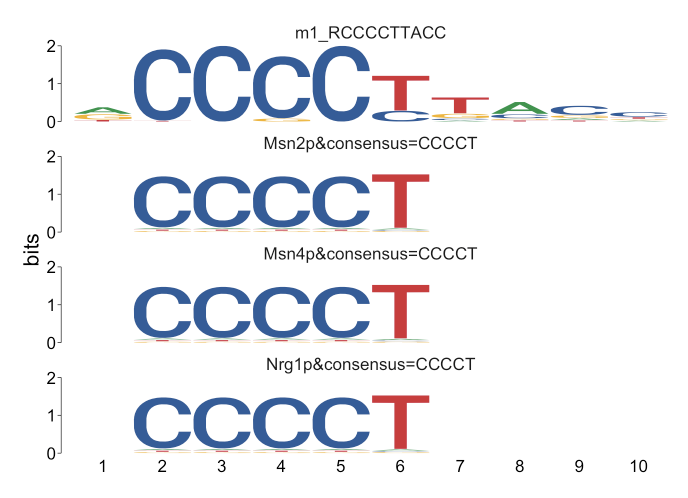
Published:
Interactive web applications for learning new alphabet scripts 
Published:
An R Package for Visualizing Differential Paralog Expression in KEGG Gene Pathways 1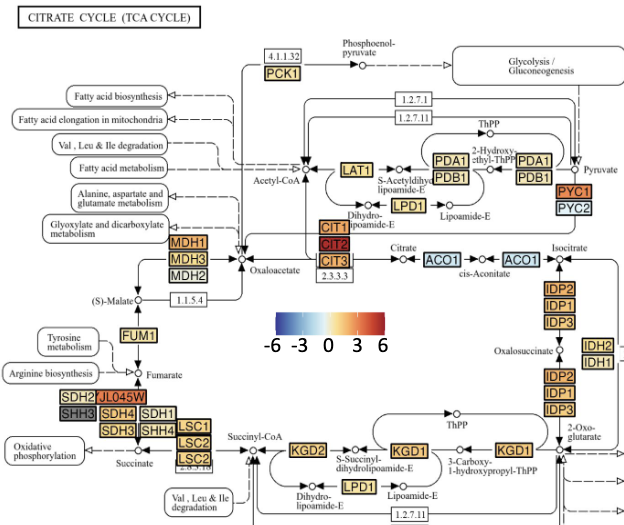
Published:
An automated grading system that saves over 100 hours per semester 
Published:
Analyze Far UV Circular Dichroism data reproducibly 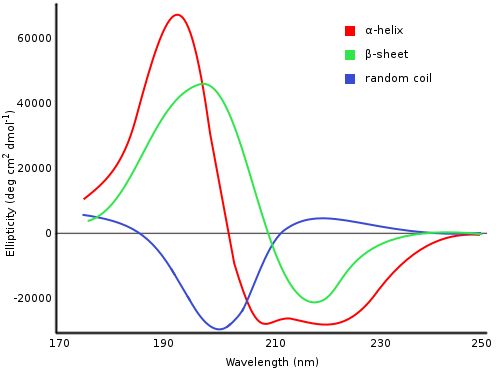
Published:
A searchable database for doubling times of haploid lab yeast knock-out strains 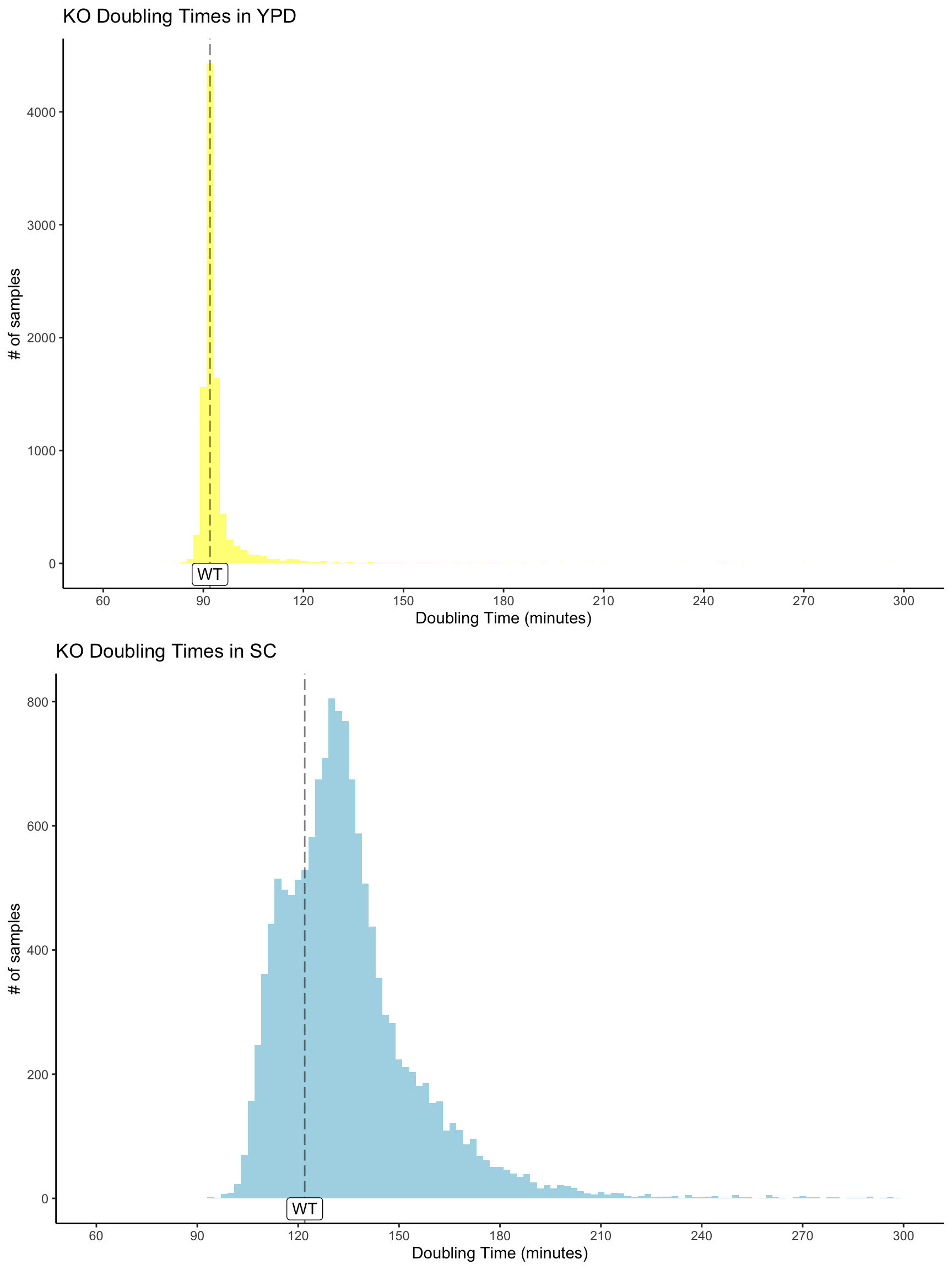
Published:
An updated Python 3 implementation of the MULTIPOOL mapping software for BSA analyses 
Published in Frontiers in Microbiology, 2020
In this paper, I quantified differential viral pathogen loads as determined by ESI/MS spectra assays and modelled inferred infection dynamics.
Recommended citation: Ceballos, Ruben Michael, Coyne Gareth Drummond, Carson Len Stacy, Elizabeth Padilla-Crespo, and Kenneth Mark Stedman. "Host-dependent differences in replication strategy of the Sulfolobus spindle-shaped virus strain SSV9 (aka, SSVK1): infection profiles in hosts of the family Sulfolobaceae." Frontiers in Microbiology 11 (2020): 1218. https://doi.org/10.3389/fmicb.2020.01218
Published in Journal of General Virology, 2021
This paper is about a temporally weighted metric of differences in growth inhibition resulting from viral infection in microbial populations.
Recommended citation: Ceballos, Ruben Michael, and Carson Len Stacy. "Quantifying relative virulence: when μ max fails and AUC alone just is not enough." Journal of General Virology 102, no. 1 (2021): 001515. [https://doi.org/10.1099/jgv.0.001515](https://doi.org/10.1099/jgv.0.001515)
Published in BMC Biology, 2024
In this paper, I introduce ordinal logistic regression of yeast survival data for analyzing changes in acquired stress resistatnce
Recommended citation: Scholes, Amanda N., Tara N. Stuecker, Stephanie E. Hood, Cader J. Locke, Carson L. Stacy, Qingyang Zhang, and Jeffrey A. Lewis. "Natural variation in yeast reveals multiple paths for acquiring higher stress resistance." BMC biology 22, no. 1 (2024): 149. https://doi.org/10.1186/s12915-024-01945-7
Published in Microorganisms, 2024
In this paper, I developed an analytical workflow and subsequently quantified weighted spectral differences between natural and engineeringed heat shock proteins.
Recommended citation: **Furr, M.**, Basha, S., Agrawal, S., Alraawi, Z., Ghosh, P., **Stacy, C. L.**, Kumar, T.K.S, & Ceballos, R. M. (2021). *Structural Stability Comparisons Between Natural and Engineered Group II Chaperonins: Are Crenarchaeal “Heat Shock” Proteins Also “pH Shock” Resistant?* **Microorganisms**, 12(11), 2348. hhttps://doi.org/10.3390/microorganisms12112348
Published in bioRxiv, 2025
In this paper, we examine transcriptomic the effects of diet on the immune response of canaries
Recommended citation: **Sauer, E. L.***, **Stacy, C. L.***, Perrine, W., Love, A. C., Lewis, J. A., & DuRant, S. E. (2024). *Diet driven differences in host tolerance are linked to shifts in global gene expression in a common avian host-pathogen system.* **bioRxiv, 2024-08.** *Co-first authors.* https://doi.org/10.1101/2024.08.07.607042
Published:
This is a description of your talk, which is a markdown files that can be all markdown-ified like any other post. Yay markdown!
Published:
To use Paralogs, go to visit the GitHub repository.
Undergraduate Supplemental Instruction Course, University of Arkansas, Department of Student Success, 2015
During my sophomore through senior years of college, I served as a Supplemental Instruction (SI) Leader for General Chemistry II, Honors General Chemistry II, and Fundamentals of Chemistry. This role allowed me to support students in mastering challenging chemistry concepts through collaborative learning sessions.
High School course, Kolahun High School, 2017
As a Peace Corps Volunteer in Liberia, I had the rewarding experience of teaching high school chemistry. During my service, I developed and implemented a comprehensive chemistry curriculum tailored to the needs of my students, who were eager to learn despite the challenging circumstances. My approach was hands-on and interactive, emphasizing practical experiments and real-world applications of chemical principles.
Undergraduate Laboratory Course, University of Arkansas, Department of Biological Sciences, 2022
From August 2022 to December 2023, I served as a Teaching Assistant for the Principles of Biology Lab, where I played a crucial role in guiding students through foundational biological concepts and laboratory techniques. My responsibilities spanned a range of instructional and support activities aimed at creating a dynamic and effective learning environment.
Undergraduate Laboratory Course, University of Arkansas, Department of Biological Sciences, 2024
As a Teaching Assistant for the Human Physiology Lab, I played a pivotal role in enhancing students’ understanding of complex physiological concepts through hands-on learning and interactive instruction. My responsibilities encompassed a range of tasks designed to support both students and faculty, ensuring a comprehensive and effective learning experience.
Graduate Bioinformatics Course, University of Arkansas, Department of Biological Sciences, 2025
As a Teaching Assistant for the Genomic Data Analysis Course, I support an advanced graduate curriculum that I co-developed with my graduate advisor in 2023. This course integrates theoretical insights with practical applications in high-throughput sequencing and bioinformatics, empowering students to tackle complex genomic datasets with confidence.